My recent works in three slides! Dara Science!
Some cool reseach that I have done includes: Large-scale Atomic/Molecular Massively Parallel Simulator: MPI + MD for Crystal Nucleation We used a cubic simulation cell with periodic boundary conditions and two crystal structures + water + NaCl to simulate nucleation of salt crystals in a nanopore. Existing open source code Large-scale Atomic/Molecular Massively Parallel Simulator (LAMMPS) were used. The calculations were carried out using 12 nodes of the University of Southern California High-Performance Computing Center, with each node having 16 processors. We published a nice paper in The Journal of Physical Chemistry Letters. Here is the Link to the paper.
Network Modeling
This includes a variety of algorithms and computational methods that I implemented. Fractional Brownian Motion, Image analysis, finite difference along with basics of graph theory were used to simulate fluid and other interesting physics.
Parallel implementation of Mid-point displacement (MPD) algorithm:
MPD is an interesting method for generating heightmaps for computer graphics. In the sequential implementation of the algorithm, as the
size of the system increases, the computation and required memory increase
exponentially, resulting in undesired runtime. In order to make the runtime manageable, I
adopt message passing interface (MPI) to implement a three-dimensional version of the MPD
algorithm. The MPI implementation on eight nodes achieves 71% to 89% efficiency for large
problem sizes compared with the sequential counterpart.
Simulation of evaporation and salt precipitation:
A computational framework was developed based on the finite difference scheme and flow network which involves invasion percolation to simulate physical phenomena such as drying of liquid and precipitation of solids.
Her is a paper that we published in Advances in Water Resources journal.
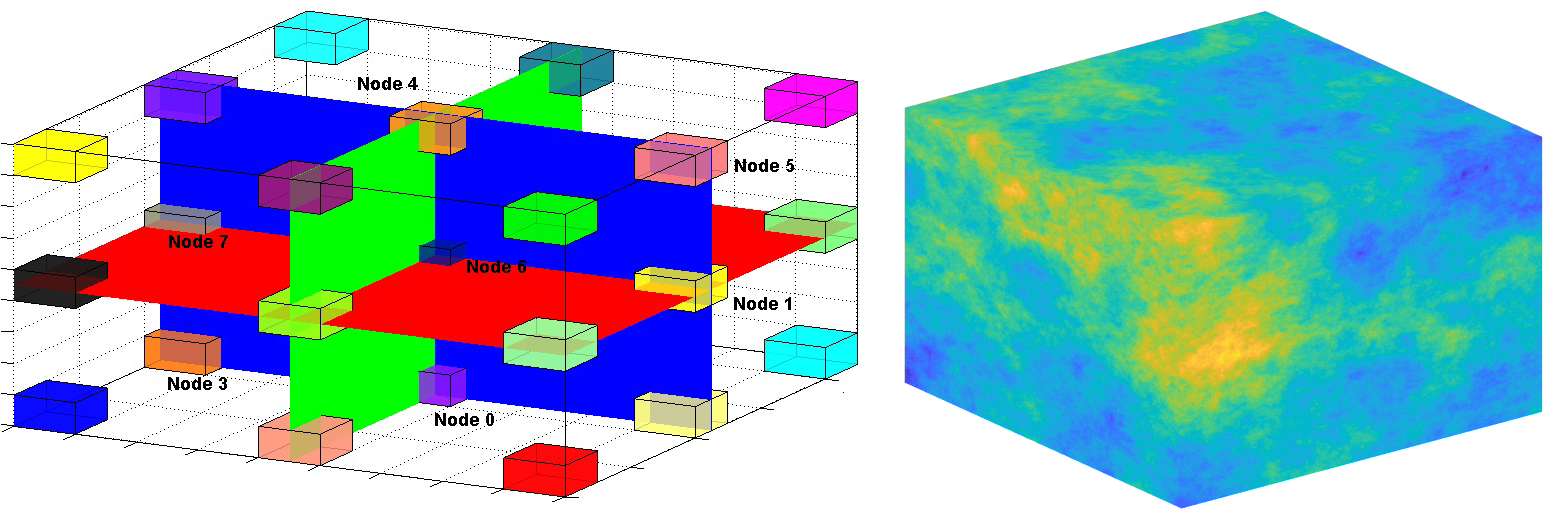
3-Dimensional flow network:
Simulation in 3D with more than 1 million nodes is computationally expensive. I did my best to optimize the computations and algorithms.

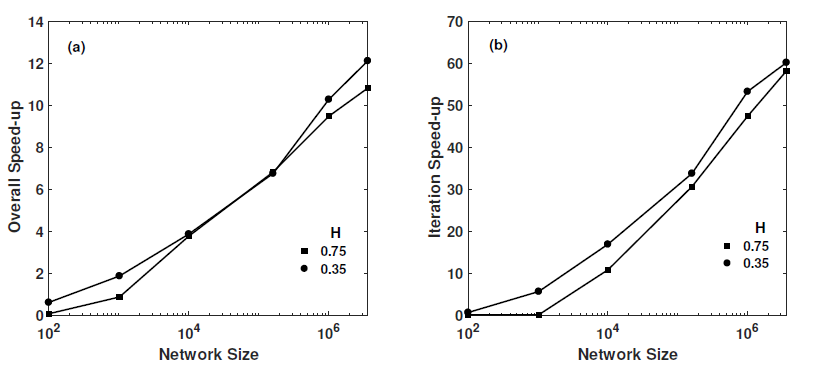
GPU based network modeling
I developed first GPU-based pore network simulator. I presented my work in GPU Technology Conference, April 4-7, Silicon Valley, CA.
It includes the MPI and CUDA to speedup calculations. Here, you can see the speedup that I got from my implementation.
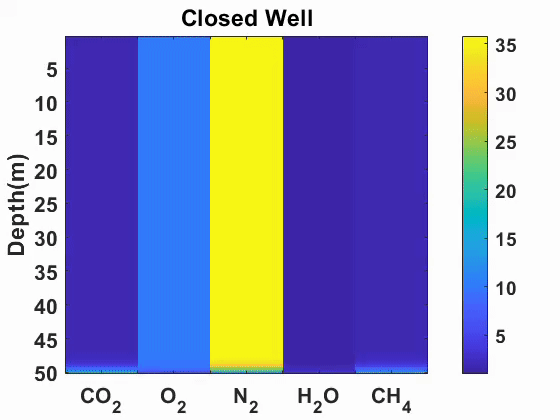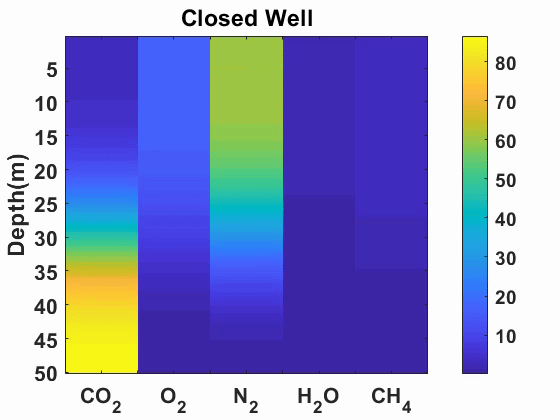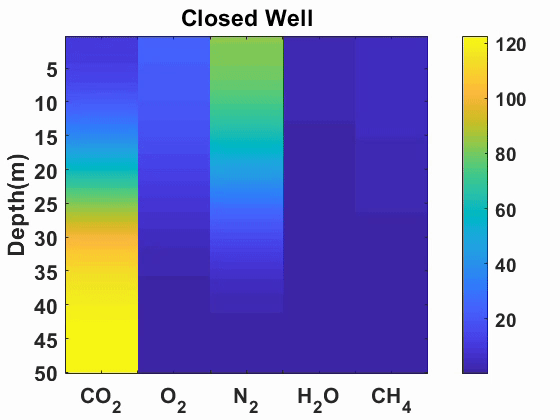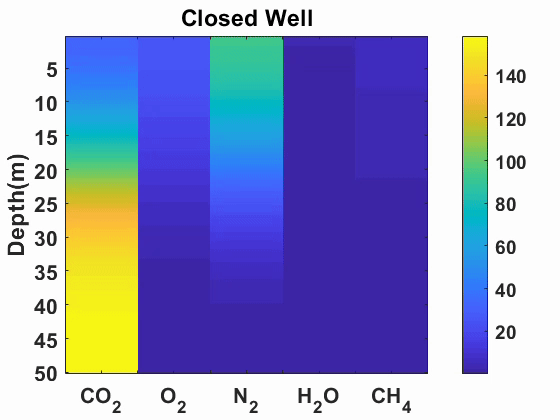
Multicomponent diffusion and advection of gases
We simulated advection and diffusion of multicomponent gases in one-dimension.
Cool Science! Compare Levy flight and Random walk Right click on each of the buttons and open in a new window to compare. When you open one in a new window, wait until your browser finishes the loading then open the other. It may take time to simulate and render, but wait! After, each window opens, you may click any where in that window to see a new simulation result. Levy Flight! Random Walk! Compare fractal images Fractional Brownian motion was used to generate 2 fractal surfaces. One with long memory and one with short memory. You can compare them here: H = 0.7 H = 0.3 Networks Here are different networks (lattices, indeed) that you can play with nodes (drag nodes with mouse) and make them smooth. 2-D Network 3-D Network Real Network Packed objects and collision Circular packed beds and rectangular objects collision are simulated here. Use your mouse to play with them. Packed Circles Packed Squares